Genomic Insights into Longevity: Shared Pathways and Unique Mechanisms in Mammals
Longevity and aging are fundamental biological processes that have intrigued scientists for decades. The variability in lifespan among different mammalian species, and even within the same species, raises questions about the underlying molecular mechanisms that control these processes. Recent research led by Alexander Tyshkovskiy and colleagues sheds light on the distinct molecular signatures associated with longevity and aging across 41 mammalian species. This groundbreaking study reveals how these signatures interact with known lifespan-extending interventions and provides insights that could lead to new therapeutic approaches for extending human healthspan and lifespan.
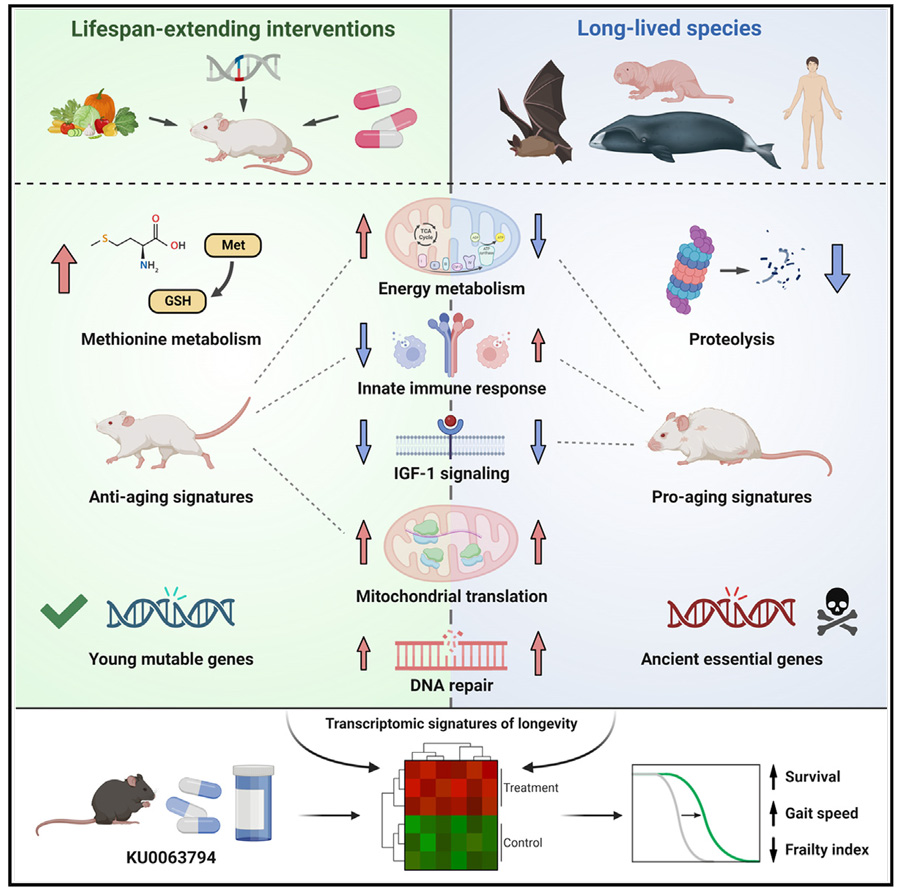
Molecular Mechanisms of Longevity
The study identified several key molecular mechanisms that regulate longevity. One of the most significant findings was the downregulation of the insulin-like growth factor 1 (Igf1) gene and the upregulation of genes involved in mitochondrial translation across long-lived species. These changes suggest that reduced IGF-1 signaling and enhanced mitochondrial function are critical components of the longevity blueprint. Additionally, the regulation of the innate immune response and cellular respiration were found to be common features among long-lived species, indicating that these processes play a vital role in lifespan regulation.
The insulin-like growth factor (IGF) pathway is well-known for its role in growth and development, and it has been extensively studied in the context of aging. The downregulation of Igf1 in long-lived species supports the idea that reduced IGF signaling can lead to increased lifespan. This is consistent with previous studies showing that mutations reducing IGF-1 signaling can extend lifespan in various organisms, including worms, flies, and mice. Mitochondrial function, another key player identified in this study, is crucial for cellular energy production. Enhanced mitochondrial translation suggests better mitochondrial health, which is known to correlate with increased lifespan. This finding aligns with the mitochondrial free radical theory of aging, which proposes that reduced mitochondrial efficiency leads to the accumulation of damaging reactive oxygen species (ROS), contributing to aging.
Biomarkers of Aging and Longevity
Aging is characterized by a range of molecular changes that occur in various tissues. The study found that these aging-related changes in gene expression were positively correlated across different tissues and species. This indicates that there are universal molecular mechanisms underlying aging. Interestingly, long-lived species exhibited gene expression profiles enriched for genes involved in proteolysis and PI3K-Akt signaling pathways, which are known to be associated with aging and longevity. These findings highlight the importance of these pathways in the regulation of lifespan.
The PI3K-Akt signaling pathway is involved in numerous cellular processes, including growth, metabolism, and survival. Dysregulation of this pathway has been linked to various age-related diseases, such as cancer and diabetes. The enrichment of PI3K-Akt signaling genes in long-lived species suggests that precise regulation of this pathway is crucial for promoting longevity. Proteolysis, the breakdown of proteins, is a vital process for maintaining cellular homeostasis. The observed changes in proteolysis-related genes imply that efficient protein turnover and degradation are essential for longevity, preventing the accumulation of damaged or misfolded proteins that can impair cellular function.
Impact of Lifespan-Extending Interventions
The study also explored how various lifespan-extending interventions impact gene expression. Interventions such as calorie restriction and growth hormone receptor knockout (GHRKO) were found to counteract the molecular changes associated with aging. These interventions primarily targeted younger, more mutable genes involved in energy metabolism, highlighting the potential of these pathways as therapeutic targets. Notably, the compound KU0063794 was identified as a potential geroprotector, extending both lifespan and healthspan in mice.
Calorie restriction (CR) is one of the most well-studied interventions known to extend lifespan across a variety of species. CR has been shown to improve metabolic health, reduce oxidative damage, and enhance stress resistance. The study's finding that CR affects younger, more mutable genes involved in energy metabolism underscores the significance of metabolic regulation in aging and longevity. The identification of KU0063794 as a lifespan-extending compound is particularly exciting. This compound was found to upregulate genes involved in mitochondrial function and downregulate those associated with inflammation and proteolysis, suggesting a comprehensive impact on several aging-related pathways.
Comprehensive Methodology
The research utilized a robust methodology, including multi-tissue RNA sequencing (RNA-seq) across 41 mammalian species. This approach allowed the researchers to identify gene expression signatures associated with longevity and to perform integrative analyses combining these signatures with known biomarkers of aging and transcriptomic data from lifespan-extending interventions. The study employed advanced statistical models, including Elastic Net linear regression, to predict lifespan based on tissue gene expression, significantly outperforming predictions based solely on body mass.
Multi-tissue RNA-seq provides a comprehensive view of gene expression across different organs, revealing tissue-specific and universal patterns. This method is particularly powerful in identifying the molecular underpinnings of complex traits like aging and longevity. The use of Elastic Net linear regression, a regularization technique that enhances the prediction accuracy by combining the properties of both ridge regression and lasso regression, enabled the researchers to create a predictive model that accounted for a significant portion of the variation in lifespan across species. This model's success underscores the importance of tissue-specific gene expression in understanding and predicting lifespan.
Significant Results
The study's results were groundbreaking,
1. Gene Expression Analysis: RNA-seq data revealed significant influences of both organ type and species on gene expression profiles. Long-lived species such as the naked mole rat, Brandt’s bat, and bowhead whale displayed unique gene expression patterns compared to shorter-lived species.
2. Predictive Modeling: The Elastic Net linear regression model captured 78% of the total variation in lifespan across species, highlighting the predictive power of tissue gene expression.
3. Functional Insights: Functional enrichment analysis showed that genes involved in translation, base excision repair, and mitochondrial function were upregulated in long-lived species, while those involved in ubiquitin-mediated proteolysis and the TCA cycle were downregulated.
These findings provide a detailed molecular framework for understanding the genetic basis of longevity. The unique gene expression patterns observed in long-lived species highlight the evolutionary adaptations that contribute to extended lifespan. The ability of the predictive model to explain a significant portion of lifespan variation underscores the complex interplay between genetics and longevity. The functional insights gained from this study reveal critical pathways that could be targeted for therapeutic interventions to promote healthy aging.
Discussion and Implications
The study provides profound insights into the complexity of lifespan regulation. While some molecular mechanisms of longevity are shared across species, others are unique to specific lineages. The identification of biomarkers common to both aging and lifespan-extending interventions offers a powerful tool for discovering new geroprotectors. The successful lifespan extension observed with KU0063794 in mice underscores the potential of targeting these molecular pathways for therapeutic interventions.
One of the key implications of this research is the potential for developing targeted therapies that can modulate the identified longevity pathways. For instance, compounds that mimic the effects of calorie restriction or enhance mitochondrial function could be promising candidates for extending healthspan and lifespan in humans. Additionally, the study's findings could inform the development of personalized medicine approaches that take into account an individual's genetic makeup and specific molecular signatures associated with aging.
The discovery of biomarkers that are shared between aging and lifespan-extending interventions also opens up new possibilities for early detection and monitoring of aging-related changes. These biomarkers could be used to develop diagnostic tools that identify individuals at risk of accelerated aging and guide interventions to mitigate these risks.
Conclusion
This comprehensive study advances our understanding of the molecular mechanisms that regulate lifespan across mammals. By identifying both universal and species-specific longevity signatures, the research opens new avenues for developing interventions aimed at extending healthspan and lifespan in humans and other animals. The discovery of compounds like KU0063794 highlights the potential for targeted therapies that leverage these molecular insights to promote longevity.
This study represents a significant step forward in the field of aging research, providing valuable data and insights that could lead to breakthroughs in extending human healthspan and lifespan. As our understanding of the molecular mechanisms underlying aging and longevity continues to grow, the possibility of developing effective anti-aging therapies becomes increasingly tangible.
The integration of multi-tissue RNA-seq data with advanced predictive modeling and functional enrichment analyses sets a new standard for research in this field. Future studies building on this foundation will likely uncover even more detailed and nuanced mechanisms of aging and longevity, paving the way for innovative approaches to enhance human health and lifespan.
References
- Tyshkovskiy, A., Ma, S., Shindyapina, A. V., et al. (2023). Distinct longevity mechanisms across and within species and their association with aging. *Cell, 186*(2929-2949). https://www.cell.com/action/showPdf?pii=S0092-8674%2823%2900476-2
By providing a detailed exploration of the distinct and shared mechanisms of longevity across different species, this study not only enhances our understanding of aging but also offers practical insights that could translate into tangible health benefits. As we continue to explore the complex interplay between genetics, environment, and aging, the potential to unlock the secrets of a longer, healthier life becomes ever more attainable.
















 English (United States) ·
English (United States) ·